|
BY JEAN CHEN David Van Essen is the winner of the prestigious 2017 Glass Brain Award from the Organization of Human Brain Mapping (OHBM). David is the Alumni Endowed Professor in the Department of Neuroscience at Washington University in St. Louis, and he chaired the department for 20 years. He was Principal Investigator (PI) for the original Young Adult Human Connectome Project (HCP, jointly with Dr. Kamil Ugurbil from the University of Minnesota) and is currently a co-PI for the Lifespan HCP Development, Lifespan HCP Aging, and Connectome Coordination Facility projects. He was also founding chair of the OHBM Council. The Glass Brain Award is selected annually "to recognize lifetime achievement by leading researchers using or facilitating neuroimaging to discover original and influential findings". David was recognized for his illustrious career of charting the brain over more than 50 years. His journey in brain mapping has taken him from Harvard to Oslo to University College London (UCL, as a postdoctoral fellow), and then as faculty at Caltech and at Wash U.
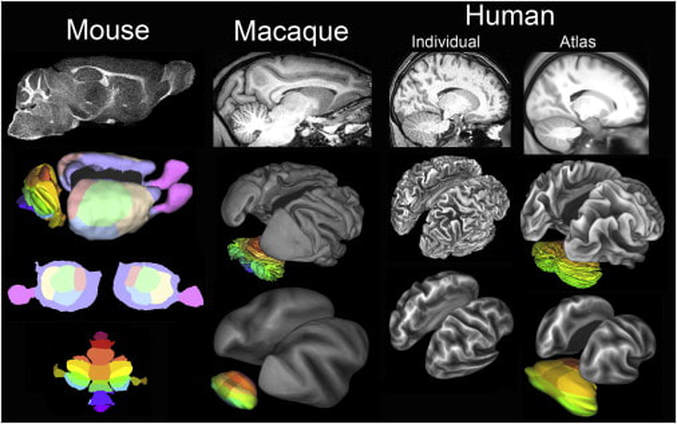
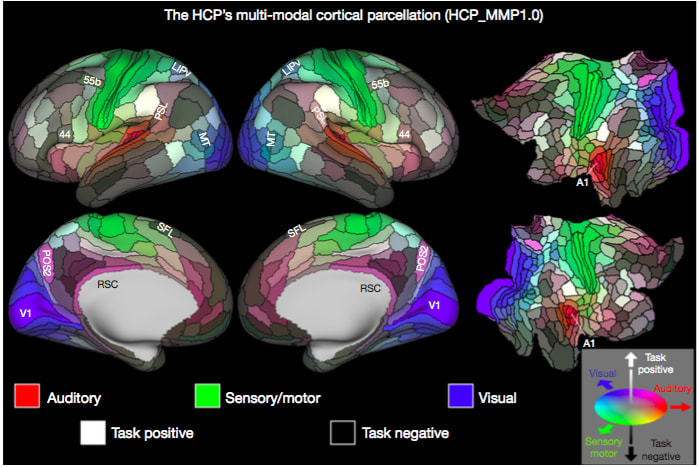
Jean Chen (JC): When and how did you begin as a “brain mapper”? David Van Essen (DVE): My adventures in brain mapping began in 1975, when I was studying extrastriate visual cortex in the macaque monkey as a postdoc at UCL. To deal with the complexity of cortical convolutions, I became a “cortical cartographer” by developing a pencil-and-tracing-paper method of making cortical flat maps, akin to flat maps of the earth’s surface. Later, my lab and others were able to computerize this process, thereby ushering in the modern era of computerized cortical cartography. JC: What do you consider your proudest academic/career achievements (aside from the Glass Brain Award)? DVE: On the scientific front, I consider my top four achievements to be: 1) Proposing that cerebral cortex is a distributed hierarchical system based on patterns of anatomical connectivity (with John Maunsell and Dan Felleman) 2) Hypothesizing that mechanical tension along axons and dendrites is a fundamental driving force for brain morphogenesis, including cortical folding. 3) Leading (with Kamil Ugurbil) the Human Connectome Project and helping develop and expound the ‘HCP-style’ neuroimaging paradigm for data acquisition, analysis, and sharing. 4) Mapping (with Matt Glasser) a new 180-area-per-hemisphere multimodal human cortical parcellation. JC: As PI of the Human Connectome Project, how did your involvement begin, and what do you see for the outcome and future of the HCP? DVE: In 2009, NIH announced a competition for the Human Connectome Project, triggering a flurry of discussions at Washington University (‘WashU’) and at many other institutions. I emerged as the leader of the WashU effort, and we later decided to team up with the University of Minnesota, Oxford University, and several other institutions to establish a consortium with broad and complementary strengths. Once the HCP was awarded in 2010, helping to lead this project became my primary research activity, and it emerged as the most exciting adventure of my scientific career. The success of the HCP can be measured in a variety of ways; e.g., nearly 10,000 investigators have agreed to HCP data use terms; more than 400 publications acknowledge HCP data use; and these numbers continue to grow. The original ‘young adult HCP’ wrapped up in 2016. It has been supplanted by a three-pronged effort, all predicated on HCP-style neuroimaging. (1) NIH awarded three Lifespan Human Connectome Projects to elucidate brain circuitry across the lifespan, during healthy development, maturation, and aging; I am one of the Principal Investigators for the Lifespan HCP Development and Aging projects. These projects are complemented by the Developing Human Connectome Project (dHCP) in Britain, which is studying brain development prenatally and at birth. (2) NIH has also funded 14 projects under the Connectomes Related to Human Disease (“Disease Connectome”) umbrella; each of these projects studies brain circuitry in a particular brain disorder. (3) All of the data from the Lifespan projects and the Disease Connectome Projects will be freely shared via the Connectome Coordination Facility (CCF) that Dan Marcus and I jointly lead. JC: In 2017, you were elected to the prestigious National Academy of Sciences. Can you describe for us how it came about and how it will influence your research going forward? DVE: One morning last May, I received an unexpected phone call while working at home, with the terrific news about my election to the NAS. The news spread quickly, and I was soon enjoying a veritable blizzard of congratulatory emails. The actual induction ceremony will be in the spring of 2018. While it is deeply gratifying to receive acknowledgment for lifetime accomplishments, I don’t anticipate that Academy membership will strongly impact my research focus or agenda. I still receive my greatest enjoyment from working in the scientific trenches with students, staff, and collaborators. JC: Your 2016 publication in Nature (“A multi-modal parcellation of human cerebral cortex”) generated much excitement. This parcellation was generated based on cortical structure, function, connectivity and topography, and identified 97 new brain areas. What’s the next step in this line of research? DVE: This study is truly a highlight of my scientific career, but the lion’s share of the credit goes to Matt Glasser (my grad student at the time) for his vision and tenacity in driving the project to fruition. Of particular importance is that our ‘areal classifier’ approach allows parcellation of individual subjects, as long as a sufficient amount of high-quality multimodal imaging data has been acquired. Several interesting next steps spring to mind. Applying the HCP multimodal parcellation strategy (using the areal classifier) to subjects from the Lifespan HCP Development and Aging projects should reveal whether some cortical areas get larger, smaller, or change their connectivity with maturation and/or aging. Applying the same approach to the Disease Connectome datasets will hopefully reveal areal differences related to brain disorders, and could potentially serve as valuable disease-specific biomarkers. We also hope that investigators in a variety of other arenas use our freely available multimodal parcellation (and the areal classifier, once it is publicly released) to aid in their own research projects by more accurately localizing and analyzing various phenomena and regions of interest. Finally, I am hopeful that the HCP multimodal parcellation and associated connectivity-related data will enable an important test of my 1997 hypothesis that axonal tension drives cortical folding. The key question is whether folding patterns in individual HCP subjects can be predicted by an analysis of ‘parcellated connectivity’. JC: You’re considered an “activist” for data sharing. How did this passion begin, and what role did you think large-scale data sharing would play in brain research?
DVE: I became interested in data sharing in 1989-90, when I served on an Institute of Medicine committee that generated a report “Towards a National Neural Circuitry Database” and helped launch the “original” Human Brain Project in 1993. The HBP was led by two visionaries, Steve Koslow and Mike Huerta, and it emphasized data sharing from the outset. My Human Brain Project grant was first funded in 1994 and is now in its 24th year, with a sustained focus on informatics tools and data sharing. Data sharing has been spectacularly successful and vital in genomics, proteomics, and other molecularly oriented domains of bioinformatics. In contrast, systems neuroscientists, and neuroimagers in particular, have been slow out of the neuroinformatics gate. This is finally changing, thanks in part to the success of several large-scale neuroimaging projects, including the HCP and OpenfMRI, that focus on sharing unprocessed data but also (especially for the HCP) ‘minimally preprocessed’ data. Sharing of large-scale neuroimaging datasets is just the tip of an important iceberg. Another important objective is to facilitate sharing of extensively processed data, such as data associated with published figures. To address the challenge of organizing and sharing complex datasets, our neuroimaging visualization and analysis software, Connectome Workbench, uses ‘scene files’ to store all of the information needed to replicate exactly what is displayed in published figures. Investigators can upload scene files and their associated datasets to the BALSA database developed by my lab. To see BALSA in action, simply click this link. JC: What do you foresee as a next “big thing” in brain mapping? An exciting new research direction or development? DVE:It’s important to note that many “big things” consist of many small or medium advances that work well in combination. For example, the success of the HCP stems from numerous advances in data acquisition, analysis, and sharing, only a few of which were ‘big’ (such as “multiband” imaging for fMRI and diffusion imaging). I hope that one big thing in human neuroimaging over the next few years will be an accelerated transition, in which a large majority of investigators adopt the best available among existing approaches to data acquisition, analysis and sharing rather than sticking with ‘traditional’ methods that have been shown to be sub-optimal yet still dominate the field. Beyond that, I anticipate continued exciting advances in neuroimaging methodology, some of which may be game-changers in terms of acquiring and analyzing high resolution data at the level of individual cortical layers and columns. Regarding invasive brain imaging in animal models, I stand in awe of the explosion of new methods of charting brain structure, function, connectivity, and gene expression at both the microscopic and mesoscopic level. These advances have far from run their course, and indeed are likely to accelerate through ongoing investments such as the BRAIN Initiative and major private funding sources (e.g., the Allen Institute and the Chan-Zuckerberg Initiative). JC: Despite how much we know about the brain now, what do you consider to be the biggest challenge(s) for brain mappers, especially in this age of “big data”? DVE: It seems that the more we know about the brain, the more we realize how much remains to be deciphered. The era of big data brings exciting but daunting challenge to center stage: how can we weave together truly staggering amounts of complex data at different scales in space and time. Capitalizing fully on these vast treasure troves of information will require major advances in neuroinformatics and computational neuroscience. I predict that over the next several decades neuroinformatics and computational neuroscience will be radically reshaped and in turn will have a transformative effect on our ability to understand the brain in health and disease. JC: What two pieces of advice do you have for young and aspiring scientists? DVE: First, stay close to your data! In general, many complex processing steps occur between the initially acquired data and that which can represent a publishable figure and associated quantitative analysis. For your own research projects, know exactly what happened to your data; be your own devil’s advocate regarding methodological problems and potential biases that might impact your interpretation; and temper your conclusions accordingly. When attending to the research of others, be constructively critical but as even-handed as possible. Second, gain as much depth and breadth as possible for whatever research area you’ve chosen. This is very challenging in modern neuroimaging, so it is also important to network closely with others who can share complementary expertise needed for whatever project(s) you’re working on. Fortunately, a growing on-line community and associated resources can allow everyone to participate, even those from small institutions and/or laboratories. [OK, this is one over the limit, but I can’t resist!] Follow your scientific passions! In today’s intensely competitive academic environment, one key for success is to be passionately interested in the scientific and technical issues you’re working on, so you can sustain the drive and energy to make it through the ups and downs of a research career.
0 Comments
Your comment will be posted after it is approved.
Leave a Reply. |
BLOG HOME
Archives
January 2024
|

 RSS Feed
RSS Feed